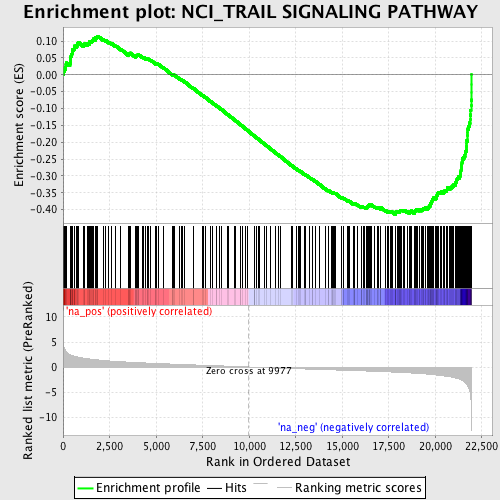
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | NCI_TRAIL SIGNALING PATHWAY |
| Enrichment Score (ES) | -0.4145497 |
| Normalized Enrichment Score (NES) | -1.9449722 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.025457771 |
| FWER p-Value | 0.22 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IL2RA | 15 | 4.747 | 0.0111 | No | ||
| 2 | NUP214 | 64 | 3.714 | 0.0181 | No | ||
| 3 | LCK | 137 | 3.203 | 0.0228 | No | ||
| 4 | PPP3CB | 153 | 3.144 | 0.0299 | No | ||
| 5 | ITCH | 197 | 2.938 | 0.0352 | No | ||
| 6 | SOS1 | 370 | 2.483 | 0.0334 | No | ||
| 7 | CSNK2A1 | 381 | 2.459 | 0.0391 | No | ||
| 8 | PRKCA | 397 | 2.430 | 0.0444 | No | ||
| 9 | CASP3 | 413 | 2.406 | 0.0497 | No | ||
| 10 | MAPKAP1 | 418 | 2.399 | 0.0555 | No | ||
| 11 | MAP3K7 | 448 | 2.353 | 0.0600 | No | ||
| 12 | AP2M1 | 478 | 2.309 | 0.0644 | No | ||
| 13 | DAPP1 | 486 | 2.292 | 0.0698 | No | ||
| 14 | CREBBP | 519 | 2.250 | 0.0739 | No | ||
| 15 | PRKDC | 584 | 2.174 | 0.0764 | No | ||
| 16 | PTEN | 596 | 2.163 | 0.0812 | No | ||
| 17 | YWHAZ | 622 | 2.135 | 0.0854 | No | ||
| 18 | RPS6KB1 | 706 | 2.046 | 0.0867 | No | ||
| 19 | IL5 | 751 | 2.009 | 0.0896 | No | ||
| 20 | GSK3B | 765 | 1.997 | 0.0940 | No | ||
| 21 | PIK3CA | 824 | 1.937 | 0.0961 | No | ||
| 22 | EIF4E | 1119 | 1.747 | 0.0869 | No | ||
| 23 | CTLA4 | 1130 | 1.742 | 0.0908 | No | ||
| 24 | MAPK8 | 1169 | 1.724 | 0.0933 | No | ||
| 25 | EEF2 | 1317 | 1.647 | 0.0906 | No | ||
| 26 | CAMK4 | 1351 | 1.628 | 0.0931 | No | ||
| 27 | SYK | 1418 | 1.598 | 0.0941 | No | ||
| 28 | PTPRC | 1419 | 1.597 | 0.0980 | No | ||
| 29 | GZMB | 1463 | 1.577 | 0.1000 | No | ||
| 30 | YWHAE | 1550 | 1.540 | 0.0998 | No | ||
| 31 | MAPK1 | 1594 | 1.523 | 0.1016 | No | ||
| 32 | TRAF6 | 1621 | 1.510 | 0.1042 | No | ||
| 33 | CSNK1A1 | 1654 | 1.498 | 0.1064 | No | ||
| 34 | ARHGEF6 | 1736 | 1.469 | 0.1064 | No | ||
| 35 | CFL2 | 1742 | 1.467 | 0.1098 | No | ||
| 36 | PAG1 | 1777 | 1.449 | 0.1118 | No | ||
| 37 | TBC1D4 | 1837 | 1.420 | 0.1126 | No | ||
| 38 | TNFSF10 | 1870 | 1.406 | 0.1146 | No | ||
| 39 | PIK3R1 | 2157 | 1.312 | 0.1047 | No | ||
| 40 | MDM2 | 2278 | 1.267 | 0.1023 | No | ||
| 41 | PTPRK | 2458 | 1.209 | 0.0970 | No | ||
| 42 | PRKCE | 2610 | 1.173 | 0.0930 | No | ||
| 43 | POU2F1 | 2833 | 1.121 | 0.0855 | No | ||
| 44 | TOP1 | 3099 | 1.061 | 0.0759 | No | ||
| 45 | RAN | 3501 | 0.968 | 0.0599 | No | ||
| 46 | HSP90AA1 | 3539 | 0.961 | 0.0605 | No | ||
| 47 | PPP3CA | 3559 | 0.957 | 0.0620 | No | ||
| 48 | IGF1 | 3560 | 0.957 | 0.0644 | No | ||
| 49 | PDPK1 | 3608 | 0.947 | 0.0646 | No | ||
| 50 | TIAM1 | 3877 | 0.895 | 0.0545 | No | ||
| 51 | PIM1 | 3908 | 0.888 | 0.0553 | No | ||
| 52 | ERC1 | 3923 | 0.885 | 0.0568 | No | ||
| 53 | AKT1S1 | 3957 | 0.880 | 0.0575 | No | ||
| 54 | SATB1 | 3979 | 0.876 | 0.0587 | No | ||
| 55 | MALT1 | 4026 | 0.868 | 0.0587 | No | ||
| 56 | CD22 | 4047 | 0.864 | 0.0600 | No | ||
| 57 | CRADD | 4268 | 0.828 | 0.0519 | No | ||
| 58 | KPNB1 | 4321 | 0.818 | 0.0515 | No | ||
| 59 | GSK3A | 4436 | 0.796 | 0.0482 | No | ||
| 60 | BAD | 4507 | 0.786 | 0.0470 | No | ||
| 61 | PDE3B | 4535 | 0.780 | 0.0476 | No | ||
| 62 | AKT3 | 4594 | 0.772 | 0.0469 | No | ||
| 63 | ETS1 | 4712 | 0.754 | 0.0434 | No | ||
| 64 | AKT1 | 4977 | 0.711 | 0.0330 | No | ||
| 65 | UBE2D3 | 4979 | 0.711 | 0.0347 | No | ||
| 66 | RASA1 | 5023 | 0.704 | 0.0344 | No | ||
| 67 | INSR | 5132 | 0.691 | 0.0312 | No | ||
| 68 | CAMK2G | 5399 | 0.649 | 0.0205 | No | ||
| 69 | EP300 | 5873 | 0.578 | 0.0001 | No | ||
| 70 | FASLG | 5895 | 0.575 | 0.0006 | No | ||
| 71 | NFATC3 | 5945 | 0.567 | -0.0003 | No | ||
| 72 | IL4 | 5986 | 0.560 | -0.0007 | No | ||
| 73 | CDKN1A | 6257 | 0.518 | -0.0119 | No | ||
| 74 | CHUK | 6349 | 0.506 | -0.0148 | No | ||
| 75 | PDGFA | 6424 | 0.496 | -0.0170 | No | ||
| 76 | BAX | 6537 | 0.482 | -0.0210 | No | ||
| 77 | CDKN2A | 6992 | 0.421 | -0.0409 | No | ||
| 78 | ASAH1 | 7016 | 0.418 | -0.0409 | No | ||
| 79 | E2F1 | 7467 | 0.357 | -0.0608 | No | ||
| 80 | CYCS | 7517 | 0.351 | -0.0622 | No | ||
| 81 | XPO1 | 7628 | 0.335 | -0.0664 | No | ||
| 82 | CDK4 | 7918 | 0.298 | -0.0790 | No | ||
| 83 | FKBP8 | 8018 | 0.285 | -0.0829 | No | ||
| 84 | RPS6 | 8236 | 0.256 | -0.0922 | No | ||
| 85 | KPNA1 | 8239 | 0.256 | -0.0917 | No | ||
| 86 | PPARG | 8381 | 0.235 | -0.0976 | No | ||
| 87 | FGR | 8504 | 0.217 | -0.1027 | No | ||
| 88 | LRDD | 8832 | 0.171 | -0.1174 | No | ||
| 89 | MAP3K1 | 8906 | 0.161 | -0.1203 | No | ||
| 90 | RAP1A | 9206 | 0.119 | -0.1338 | No | ||
| 91 | TNFRSF1A | 9263 | 0.111 | -0.1361 | No | ||
| 92 | DIABLO | 9521 | 0.071 | -0.1478 | No | ||
| 93 | IL3 | 9524 | 0.071 | -0.1477 | No | ||
| 94 | TNFRSF10B | 9656 | 0.050 | -0.1537 | No | ||
| 95 | BTK | 9783 | 0.031 | -0.1594 | No | ||
| 96 | CTSD | 9890 | 0.014 | -0.1643 | No | ||
| 97 | CALM2 | 10266 | -0.045 | -0.1814 | No | ||
| 98 | EGR4 | 10291 | -0.048 | -0.1824 | No | ||
| 99 | BLNK | 10309 | -0.050 | -0.1831 | No | ||
| 100 | APAF1 | 10407 | -0.066 | -0.1874 | No | ||
| 101 | LMNB1 | 10486 | -0.076 | -0.1908 | No | ||
| 102 | CASP1 | 10532 | -0.083 | -0.1927 | No | ||
| 103 | EGF | 10831 | -0.129 | -0.2061 | No | ||
| 104 | AKAP5 | 10935 | -0.146 | -0.2105 | No | ||
| 105 | IRS1 | 11122 | -0.170 | -0.2187 | No | ||
| 106 | RAF1 | 11386 | -0.204 | -0.2303 | No | ||
| 107 | NRAS | 11559 | -0.223 | -0.2377 | No | ||
| 108 | HRAS | 11684 | -0.237 | -0.2428 | No | ||
| 109 | AP2A1 | 12279 | -0.305 | -0.2695 | No | ||
| 110 | TRAF2 | 12318 | -0.311 | -0.2704 | No | ||
| 111 | LMNB2 | 12514 | -0.335 | -0.2786 | No | ||
| 112 | SLC2A4 | 12634 | -0.349 | -0.2832 | No | ||
| 113 | PPP5C | 12697 | -0.355 | -0.2852 | No | ||
| 114 | POU2F2 | 12770 | -0.364 | -0.2876 | No | ||
| 115 | TBX21 | 12958 | -0.386 | -0.2953 | No | ||
| 116 | RHEB | 13014 | -0.393 | -0.2969 | No | ||
| 117 | BLK | 13045 | -0.396 | -0.2973 | No | ||
| 118 | MAP3K8 | 13234 | -0.417 | -0.3049 | No | ||
| 119 | KRAS | 13393 | -0.438 | -0.3111 | No | ||
| 120 | ARF5 | 13414 | -0.441 | -0.3109 | No | ||
| 121 | SMPD1 | 13543 | -0.455 | -0.3157 | No | ||
| 122 | SRC | 13765 | -0.482 | -0.3247 | No | ||
| 123 | BIRC2 | 14071 | -0.516 | -0.3375 | No | ||
| 124 | TRADD | 14236 | -0.537 | -0.3437 | No | ||
| 125 | RNF128 | 14244 | -0.538 | -0.3427 | No | ||
| 126 | CSF2 | 14405 | -0.557 | -0.3487 | No | ||
| 127 | REL | 14487 | -0.568 | -0.3510 | No | ||
| 128 | ARFIP2 | 14492 | -0.569 | -0.3498 | No | ||
| 129 | SPHK2 | 14517 | -0.572 | -0.3495 | No | ||
| 130 | YES1 | 14598 | -0.581 | -0.3517 | No | ||
| 131 | MAP2K4 | 14640 | -0.588 | -0.3522 | No | ||
| 132 | ELK1 | 14948 | -0.623 | -0.3648 | No | ||
| 133 | PLCG2 | 14956 | -0.624 | -0.3635 | No | ||
| 134 | YWHAQ | 15062 | -0.634 | -0.3668 | No | ||
| 135 | CALM1 | 15082 | -0.637 | -0.3661 | No | ||
| 136 | IFNG | 15303 | -0.665 | -0.3746 | No | ||
| 137 | DOK1 | 15319 | -0.668 | -0.3736 | No | ||
| 138 | MAPK3 | 15401 | -0.677 | -0.3757 | No | ||
| 139 | RIPK2 | 15580 | -0.698 | -0.3822 | No | ||
| 140 | SMPD3 | 15637 | -0.706 | -0.3830 | No | ||
| 141 | TSC2 | 15645 | -0.708 | -0.3816 | No | ||
| 142 | PAWR | 15663 | -0.710 | -0.3806 | No | ||
| 143 | EIF4EBP1 | 15826 | -0.732 | -0.3862 | No | ||
| 144 | PPP2R5D | 16006 | -0.758 | -0.3926 | No | ||
| 145 | MAP2K2 | 16032 | -0.762 | -0.3919 | No | ||
| 146 | FADD | 16038 | -0.763 | -0.3902 | No | ||
| 147 | PLD2 | 16134 | -0.775 | -0.3927 | No | ||
| 148 | ZAP70 | 16176 | -0.781 | -0.3926 | No | ||
| 149 | CASP8 | 16284 | -0.795 | -0.3956 | No | ||
| 150 | CFLAR | 16298 | -0.798 | -0.3942 | No | ||
| 151 | FOSL1 | 16325 | -0.800 | -0.3934 | No | ||
| 152 | INPPL1 | 16331 | -0.801 | -0.3916 | No | ||
| 153 | IBTK | 16362 | -0.806 | -0.3910 | No | ||
| 154 | CD40LG | 16375 | -0.807 | -0.3896 | No | ||
| 155 | NFKBIA | 16391 | -0.809 | -0.3882 | No | ||
| 156 | HCK | 16436 | -0.815 | -0.3882 | No | ||
| 157 | YWHAB | 16464 | -0.818 | -0.3875 | No | ||
| 158 | ARF1 | 16466 | -0.818 | -0.3855 | No | ||
| 159 | SREBF1 | 16535 | -0.827 | -0.3865 | No | ||
| 160 | CD79B | 16562 | -0.830 | -0.3857 | No | ||
| 161 | MAP3K14 | 16757 | -0.855 | -0.3925 | No | ||
| 162 | RIPK1 | 16874 | -0.871 | -0.3957 | No | ||
| 163 | CASP4 | 16907 | -0.877 | -0.3950 | No | ||
| 164 | ARF6 | 16953 | -0.884 | -0.3949 | No | ||
| 165 | PTGS2 | 17046 | -0.900 | -0.3969 | No | ||
| 166 | GATA3 | 17055 | -0.902 | -0.3950 | No | ||
| 167 | AKT2 | 17075 | -0.904 | -0.3936 | No | ||
| 168 | BIRC3 | 17339 | -0.944 | -0.4034 | No | ||
| 169 | MAPK9 | 17448 | -0.962 | -0.4060 | No | ||
| 170 | RHOA | 17480 | -0.969 | -0.4050 | No | ||
| 171 | RELA | 17566 | -0.984 | -0.4065 | No | ||
| 172 | PRKACA | 17631 | -0.996 | -0.4070 | No | ||
| 173 | YWHAH | 17687 | -1.006 | -0.4070 | No | ||
| 174 | CD19 | 17851 | -1.031 | -0.4120 | Yes | ||
| 175 | PRKRA | 17878 | -1.038 | -0.4106 | Yes | ||
| 176 | BID | 17883 | -1.039 | -0.4082 | Yes | ||
| 177 | ARFGAP1 | 17885 | -1.039 | -0.4057 | Yes | ||
| 178 | DFFA | 17956 | -1.052 | -0.4063 | Yes | ||
| 179 | PARP1 | 18005 | -1.059 | -0.4059 | Yes | ||
| 180 | DGKA | 18084 | -1.073 | -0.4068 | Yes | ||
| 181 | JUN | 18099 | -1.076 | -0.4048 | Yes | ||
| 182 | SHC1 | 18103 | -1.076 | -0.4022 | Yes | ||
| 183 | CDKN1B | 18144 | -1.082 | -0.4014 | Yes | ||
| 184 | PLCG1 | 18203 | -1.093 | -0.4013 | Yes | ||
| 185 | CASP2 | 18290 | -1.111 | -0.4026 | Yes | ||
| 186 | MKNK1 | 18366 | -1.127 | -0.4032 | Yes | ||
| 187 | EIF4A1 | 18503 | -1.154 | -0.4066 | Yes | ||
| 188 | VAV2 | 18616 | -1.178 | -0.4089 | Yes | ||
| 189 | BCL2 | 18625 | -1.179 | -0.4063 | Yes | ||
| 190 | ATM | 18685 | -1.190 | -0.4061 | Yes | ||
| 191 | EIF4G1 | 18692 | -1.193 | -0.4034 | Yes | ||
| 192 | GRB2 | 18879 | -1.237 | -0.4089 | Yes | ||
| 193 | NFKBIB | 18902 | -1.241 | -0.4068 | Yes | ||
| 194 | PLEKHA2 | 18909 | -1.243 | -0.4040 | Yes | ||
| 195 | IKBKG | 18928 | -1.246 | -0.4017 | Yes | ||
| 196 | CYLD | 18965 | -1.255 | -0.4003 | Yes | ||
| 197 | GAS2 | 19048 | -1.271 | -0.4009 | Yes | ||
| 198 | CASP9 | 19125 | -1.289 | -0.4012 | Yes | ||
| 199 | DFFB | 19145 | -1.295 | -0.3989 | Yes | ||
| 200 | PRKAB1 | 19264 | -1.327 | -0.4010 | Yes | ||
| 201 | PRKCQ | 19298 | -1.339 | -0.3992 | Yes | ||
| 202 | TNF | 19341 | -1.351 | -0.3978 | Yes | ||
| 203 | RAC1 | 19377 | -1.360 | -0.3960 | Yes | ||
| 204 | GOSR2 | 19461 | -1.383 | -0.3964 | Yes | ||
| 205 | TSC1 | 19466 | -1.385 | -0.3931 | Yes | ||
| 206 | CALM3 | 19591 | -1.421 | -0.3953 | Yes | ||
| 207 | DAP3 | 19633 | -1.434 | -0.3937 | Yes | ||
| 208 | BAG4 | 19659 | -1.443 | -0.3912 | Yes | ||
| 209 | TP53 | 19687 | -1.455 | -0.3889 | Yes | ||
| 210 | CD4 | 19717 | -1.467 | -0.3865 | Yes | ||
| 211 | TNFAIP3 | 19726 | -1.471 | -0.3833 | Yes | ||
| 212 | PTK2 | 19751 | -1.476 | -0.3807 | Yes | ||
| 213 | ITK | 19776 | -1.489 | -0.3781 | Yes | ||
| 214 | APP | 19814 | -1.503 | -0.3761 | Yes | ||
| 215 | FKBP1A | 19825 | -1.506 | -0.3728 | Yes | ||
| 216 | GGA3 | 19827 | -1.506 | -0.3691 | Yes | ||
| 217 | COPA | 19881 | -1.521 | -0.3678 | Yes | ||
| 218 | MYC | 19912 | -1.534 | -0.3653 | Yes | ||
| 219 | VAV1 | 19995 | -1.568 | -0.3652 | Yes | ||
| 220 | KSR1 | 20065 | -1.594 | -0.3644 | Yes | ||
| 221 | ACTA1 | 20070 | -1.597 | -0.3606 | Yes | ||
| 222 | SFN | 20083 | -1.603 | -0.3572 | Yes | ||
| 223 | FYN | 20107 | -1.612 | -0.3543 | Yes | ||
| 224 | IKBKB | 20132 | -1.623 | -0.3513 | Yes | ||
| 225 | CASP6 | 20177 | -1.642 | -0.3493 | Yes | ||
| 226 | PPP3CC | 20266 | -1.683 | -0.3492 | Yes | ||
| 227 | PRKCZ | 20308 | -1.703 | -0.3468 | Yes | ||
| 228 | SLC3A2 | 20423 | -1.750 | -0.3477 | Yes | ||
| 229 | MAP2K1 | 20473 | -1.779 | -0.3456 | Yes | ||
| 230 | PRKCD | 20514 | -1.805 | -0.3429 | Yes | ||
| 231 | LIMK1 | 20601 | -1.848 | -0.3423 | Yes | ||
| 232 | FRAP1 | 20627 | -1.863 | -0.3388 | Yes | ||
| 233 | MAF | 20636 | -1.867 | -0.3346 | Yes | ||
| 234 | PTPN1 | 20769 | -1.947 | -0.3358 | Yes | ||
| 235 | CLTA | 20840 | -1.992 | -0.3341 | Yes | ||
| 236 | MEF2D | 20891 | -2.028 | -0.3314 | Yes | ||
| 237 | NFKB1 | 20944 | -2.065 | -0.3286 | Yes | ||
| 238 | FBXW11 | 20996 | -2.114 | -0.3257 | Yes | ||
| 239 | CLTB | 21066 | -2.158 | -0.3235 | Yes | ||
| 240 | NUMA1 | 21077 | -2.170 | -0.3186 | Yes | ||
| 241 | MAPK14 | 21120 | -2.209 | -0.3150 | Yes | ||
| 242 | BCL10 | 21159 | -2.247 | -0.3112 | Yes | ||
| 243 | ARHGEF7 | 21179 | -2.271 | -0.3064 | Yes | ||
| 244 | ARRB2 | 21227 | -2.318 | -0.3028 | Yes | ||
| 245 | FOXP3 | 21314 | -2.435 | -0.3008 | Yes | ||
| 246 | CARD11 | 21330 | -2.461 | -0.2953 | Yes | ||
| 247 | PRF1 | 21361 | -2.499 | -0.2905 | Yes | ||
| 248 | GBF1 | 21377 | -2.528 | -0.2849 | Yes | ||
| 249 | PLEKHA1 | 21384 | -2.541 | -0.2789 | Yes | ||
| 250 | ARHGDIB | 21394 | -2.566 | -0.2729 | Yes | ||
| 251 | CD72 | 21419 | -2.609 | -0.2675 | Yes | ||
| 252 | SH3BP5 | 21431 | -2.642 | -0.2615 | Yes | ||
| 253 | JUNB | 21440 | -2.670 | -0.2552 | Yes | ||
| 254 | CBLB | 21453 | -2.694 | -0.2491 | Yes | ||
| 255 | GSN | 21499 | -2.780 | -0.2442 | Yes | ||
| 256 | CASP7 | 21563 | -2.954 | -0.2398 | Yes | ||
| 257 | PTPN6 | 21597 | -3.085 | -0.2336 | Yes | ||
| 258 | NFKB2 | 21615 | -3.137 | -0.2266 | Yes | ||
| 259 | CD79A | 21671 | -3.365 | -0.2208 | Yes | ||
| 260 | YWHAG | 21678 | -3.403 | -0.2126 | Yes | ||
| 261 | FCGR2B | 21689 | -3.450 | -0.2045 | Yes | ||
| 262 | CSK | 21693 | -3.473 | -0.1960 | Yes | ||
| 263 | PRKCH | 21736 | -3.724 | -0.1887 | Yes | ||
| 264 | NFATC2 | 21737 | -3.736 | -0.1794 | Yes | ||
| 265 | MAP4K4 | 21744 | -3.794 | -0.1702 | Yes | ||
| 266 | RELB | 21749 | -3.826 | -0.1609 | Yes | ||
| 267 | IRF4 | 21795 | -4.216 | -0.1525 | Yes | ||
| 268 | LMNA | 21836 | -4.610 | -0.1429 | Yes | ||
| 269 | BCL3 | 21865 | -4.977 | -0.1318 | Yes | ||
| 270 | NFATC1 | 21871 | -5.174 | -0.1192 | Yes | ||
| 271 | VIM | 21890 | -5.636 | -0.1060 | Yes | ||
| 272 | FOS | 21917 | -6.478 | -0.0911 | Yes | ||
| 273 | NR4A1 | 21923 | -6.633 | -0.0748 | Yes | ||
| 274 | EGR1 | 21942 | -8.847 | -0.0537 | Yes | ||
| 275 | EGR3 | 21943 | -9.744 | -0.0294 | Yes | ||
| 276 | EGR2 | 21944 | -11.902 | 0.0001 | Yes |
